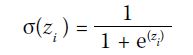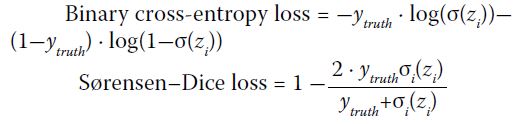© Hong Kong Academy of Medicine. CC BY-NC-ND 4.0
ORIGINAL ARTICLE CME
Artificial intelligence for detection of intracranial haemorrhage on head computed tomography scans: diagnostic accuracy in Hong Kong
Jill M Abrigo, MD, FRCR; Ka-long Ko, MPhil; Qianyun Chen, MSc; Billy MH Lai, MB, BS, FHKAM (Radiology); Tom CY Cheung, MB ChB, FHKAM (Radiology); Winnie CW Chu, MB ChB, FHKAM (Radiology); Simon CH Yu, MB, BS, FHKAM (Radiology)
Department of Imaging and Interventional Radiology, Prince of Wales Hospital, The Chinese University of Hong Kong, Hong Kong SAR, China
Corresponding author: Dr Jill M Abrigo (jillabrigo@cuhk.edu.hk)
Abstract
Introduction: The use of artificial intelligence (AI)
to identify acute intracranial haemorrhage (ICH)
on computed tomography (CT) scans may facilitate
initial imaging interpretation in the accident
and emergency department. However, AI model
construction requires a large amount of annotated
data for training, and validation with real-world
data has been limited. We developed an algorithm
using an open-access dataset of CT slices, then
assessed its utility in clinical practice by validating
its performance on CT scans from our institution.
Methods: Using a publicly available international
dataset of >750 000 expert-labelled CT slices, we
developed an AI model which determines ICH
probability for each CT scan and nominates five
potential ICH-positive CT slices for review. We
validated the model using retrospective data from
1372 non-contrast head CT scans (84 [6.1%] with
ICH) collected at our institution.
Results: The model achieved an area under the
curve of 0.842 (95% confidence interval=0.791-0.894;
P<0.001) for scan-based detection of ICH. A pre-specified
probability threshold of ≥50% for the
presence of ICH yielded 78.6% accuracy, 73% sensitivity, 79% specificity, 18.6% positive predictive
value, and 97.8% negative predictive value. There
were 62 true-positive scans and 22 false-negative
scans, which could be reduced to six false-negative
scans by manual review of model-nominated CT
slices.
Conclusions: Our model exhibited good accuracy in the CT scan–based detection of ICH, considering
the low prevalence of ICH in Hong Kong. Model
refinement to allow direct localisation of ICH will
facilitate the use of AI solutions in clinical practice.
New knowledge added by this study
- A deep learning–based artificial intelligence model trained on an international dataset of computed tomography (CT) slices exhibited good accuracy in the detection of intracranial haemorrhage (ICH) on CT scans in Hong Kong.
- Considering the 6% prevalence of ICH in our institution, and using a pre-specified probability threshold of ≥50%, the model detected 74% of ICH-positive scans; this outcome improved to 93% via manual review of model-nominated images.
- Considering the expected clinical applications, model refinement is needed to improve diagnostic performance prior to additional tests in a clinical setting.
- Our model may facilitate assessment of CT scans by physicians with different degrees of experience in ICH detection, an important aspect of real-world clinical practice.
Introduction
Head computed tomography (CT) scans constitute
the main imaging investigation during the evaluation
of trauma and stroke; they are also important in the
initial work-up of headache and other non-specific
neurological complaints. In Prince of Wales Hospital
of Hong Kong alone, >25 000 head CT scans were
performed in 2019 during the clinical management of patients who presented to the Accident and
Emergency Department. Computed tomography
scans are composed of multiple cross-sectional
images (ie, slices), which may be challenging to
interpret. Typically, these scans are initially reviewed
by frontline physicians prior to assessment by
radiologists, and delays during the review process
can be substantial. Thus, the timely recognition of an acute finding, such as intracranial haemorrhage
(ICH), is limited by the competence and availability
of frontline physicians.
The presence and location or type of ICH
impacts the next clinical step, which can be further
imaging investigations, medical management, or
surgical intervention.1 Furthermore, a confirmation
of ICH absence can also be useful in clinical
management. For example, it can facilitate safe
discharge from the hospital when appropriate; in
patients with acute stroke, the absence of ICH is
an important exclusion criterion that influences
treatment selection.2
The use of artificial intelligence (AI) for ICH
detection is a topic with global relevance considering
its diagnostic impact and ability to optimise
workflow, both of which have high practical value.3 4
In the accident and emergency department, AI can
facilitate ICH detection in head CT scans during
times when a radiologist is unavailable. Although
there have been multiple reports of deep learning
methods with high accuracy in the detection of ICH,
the models in those reports were developed using
in-house labelled training datasets and validated
using a limited number of cases.3 5 6 7 8 Recently, the
Radiological Society of North America (RSNA)
publicly released >25 000 multi-centre head CT scans
with slices that have been labelled with or without
ICH by experts.9 Here, we developed a model using
this RSNA dataset, then validated its performance
on CT scans from our institution to determine its
potential for clinical application in Hong Kong.
Methods
Ethical considerations
This study was approved by the Joint Chinese
University of Hong Kong—New Territories East
Cluster Clinical Research Ethics Committee (Ref No.: 2020.061). The model was developed from a publicly
available dataset and validated on retrospectively
acquired data from our institution. The requirement
for patient consent was waived by the Committee
given the retrospective design of the study and
anonymisation of all CT scans prior to use.
The results of this diagnostic accuracy study
are reported in accordance with the Standards
for Reporting of Diagnostic Accuracy Studies
guidelines.10
Public dataset: model development and
internal validation
We acquired 25 312 head CT scans from four
institutions in North and South America available in
the RNSA open dataset,11 and were split into slices
(each slice ≥5 mm thick), which were then randomly
shuffled and annotated by 60 volunteer experts from
the American Society of Neuroradiology. Each CT slice was labelled to indicate the presence and type of
ICH. When present, ICH was classified according to
its location, namely, intraparenchymal haemorrhage
(IPH), subarachnoid haemorrhage (SAH), subdural
haemorrhage (SDH), epidural haemorrhage (EDH),
and intraventricular haemorrhage (IVH). The RSNA
dataset comprised 752 807 CT slices, which were
divided into a training subset (85%) and test subset
(15%) for internal validation. Each subset consisted
of approximately 86% negative ICH slices and
14% positive ICH slices, along with the following
proportions of ICH subtypes: 4.8% IPH, 4.7%-4.8%
SAH, 6.3% SDH, 0.4% EDH, and 3.4%-3.5% IVH.
The convolutional neural network (CNN)
VGG (named after the Visual Geometry Group from
the University of Oxford, United Kingdom) is an
effective end-to-end algorithm for image detection.12
In this study, we adopted the VGG architecture
with a customised output layer and loss function
optimised for multi-label classification. To adjust
for the low prevalence of ICH in the training set,
each subtype’s logit outputs zi were concatenated as
independent channels after a sigmoid output layer:
The performance of the CNN model was
evaluated by binary cross-entropy loss and
Sørensen–Dice loss13:
The loss functions were linearly combined
with weighted values to produce the multi-label
classification loss:
Where wi denotes the class prevalence weight, and α and β denote respective loss mix ratios. For
simplicity, wi=1∕(n-1) for all subtype classes and wi=1 for ‘ANY’ was treated as an independent ICH
class.
The model was trained with software written
in our laboratory using the end-to-end open-source
machine learning platform TensorFlow on an Nvidia
Titan Xp graphics processing unit.
During internal validation (ie, slice-level
performance for the detection of any type of ICH),
the model achieved an area under the curve (AUC)
of 0.912 (95% confidence interval [CI]=0.909-0.915) with sensitivity and specificity of 85% and 80%, respectively. Additionally, for the detection of
specific types of ICH, the following AUC (95% CI)
and sensitivity/specificity values were obtained:
0.860 (0.853-0.867) and 77%/88% for IPH, 0.835
(0.829-0.842) and 75%/82% for SAH, 0.850 (0.845-0.855) and 74%/83% for SDH, 0.813 (0.790-0.836)
and 72%/80% for EDH, and 0.870 (0.861-0.879) and
79%/89% for IVH.
Prince of Wales Hospital dataset: external
validation
The consecutive head CT scans of patients aged ≥18
years who underwent initial brain CT scans in the
Accident and Emergency Department of Prince of
Wales Hospital from 1 to 31 July 2019 were included,
thereby simulating the point prevalence of ICH.
Head CT scans were acquired on a 64-slice
CT scanner. Data analyses were conducted using
reformatted 5-mm-thick slices, which can be
accessed and viewed by physicians at all hospital
workstations. DICOM (Digital Imaging and
Communications in Medicine) images were de-identified
prior to data analyses. The large volume
of data was explored through the identification of
relevant CT data using an automated program which
selected scans with specific DICOM tags. Computed
tomography scans performed for follow-up purposes
or after recent intracranial surgery, as well as scans
without radiologist reports, were excluded from the
analysis.
We reviewed the corresponding radiology
reports to determine the presence and type of ICH
(IPH, SAH, SDH, EDH, or IVH). The CT scans were
assessed by radiologists or senior radiology trainees;
the corresponding reports were regarded as scan-level ground truth labels for analysis, consistent with
their use as clinical reference standards in Hong
Kong. Considering its rarity, EDH was grouped with
and labelled as SDH, which has a similar appearance
on CT. For scans with false-negative results, we
performed post-hoc labelling of model-nominated
CT slices. All scans were assessed prior to model
construction; thus, the scan reports were established
without knowledge of the AI results. Furthermore,
all scans comprised the external validation dataset
and constituted ‘unseen data’ for the model.
Statistical analysis
The diagnostic accuracies of the model for the
detection of any type of ICH and each type of ICH
were determined by calculation of the AUC with
95% CI, using DeLong et al’s method.14 To construct
the confusion matrix during external validation,
CT scans were classified as ICH-positive using
a pre-specified probability threshold of ≥50%8;
the corresponding sensitivity, specificity, positive
predictive value (PPV), negative predictive value
(NPV), and accuracy were calculated. Additional
probability thresholds were established to achieve
90% sensitivity and 90% specificity for the presence
of any type of ICH. Statistical analysis was performed
using R software (version 4.0.2; R Foundation for
Statistical Computing, Vienna, Austria), and the
threshold for statistical significance was set at
P<0.05.
Results
Model output
Figure 1 shows an example of the model output. The
model report includes an overall probability for the
presence of ICH (labelled ‘A’ in Fig 1). Additionally,
the model selects five representative CT image slices
which are likely to contain ICH (one such slice is
labelled ‘B’ in Fig 1), along with the probability of
each ICH type in each representative slice (labelled
‘C’ in Fig 1). All scans were successfully analysed by
the model.
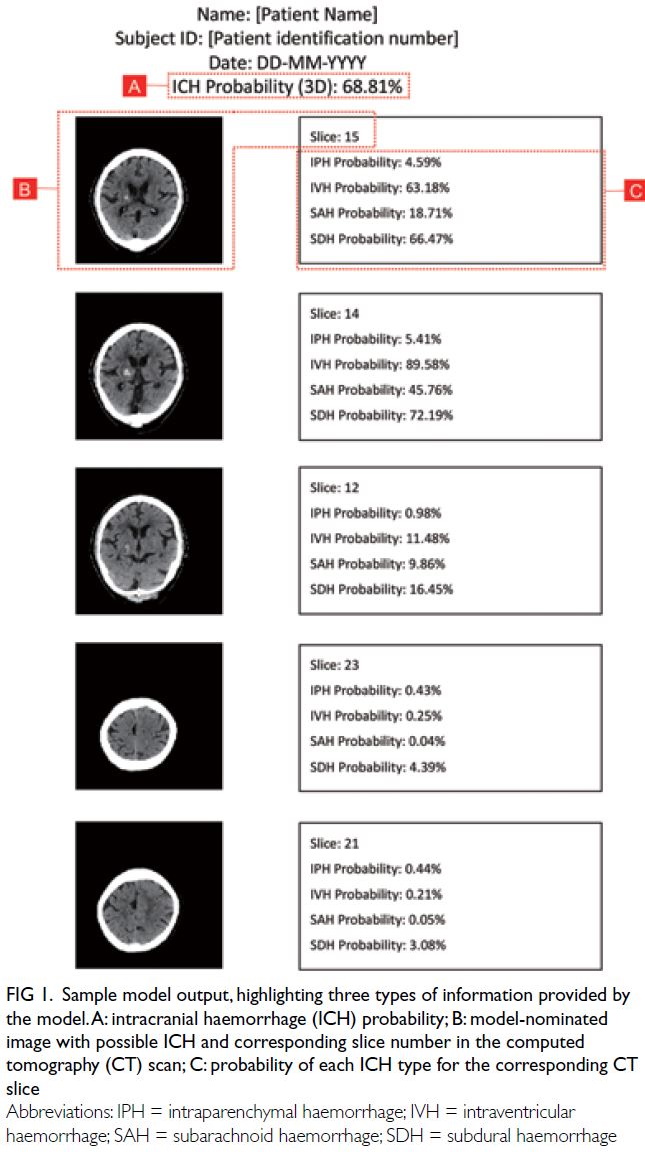
Figure 1. Sample model output, highlighting three types of information provided by the model. A: intracranial haemorrhage (ICH) probability; B: model-nominated image with possible ICH and corresponding slice number in the computed tomography (CT) scan; C: probability of each ICH type for the corresponding CT slice
Prince of Wales Hospital data and model
validation
The Standards for Reporting of Diagnostic Accuracy
Studies diagram and corresponding confusion matrix
are shown in Figure 2. In total, 1372 head CT scans
(84 [6.1%] with ICH) were included in the analysis.
The distribution of ICH types is summarised in the
Table.
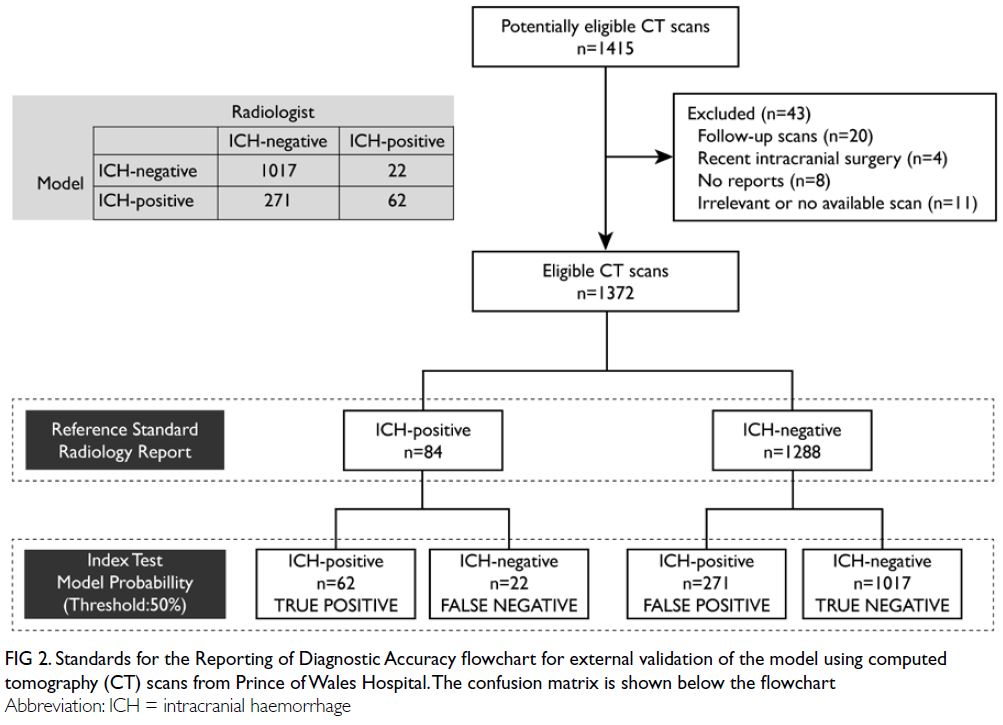
Figure 2. Standards for the Reporting of Diagnostic Accuracy flowchart for external validation of the model using computed tomography (CT) scans from Prince of Wales Hospital. The confusion matrix is shown below the flowchart
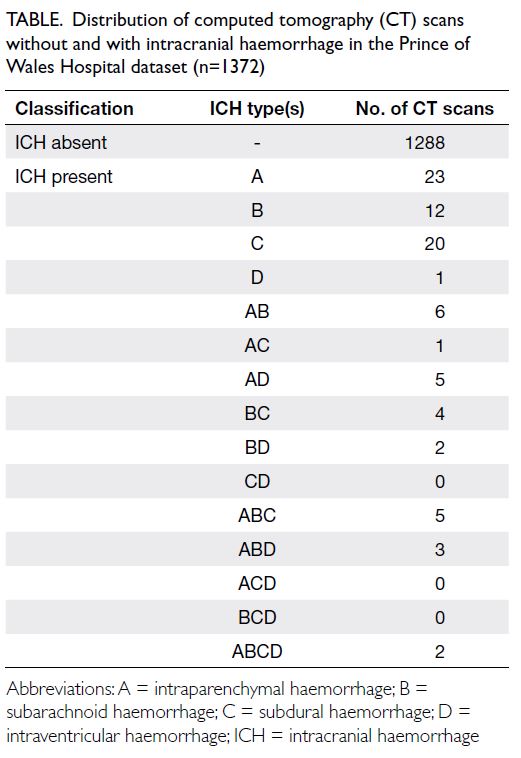
Table. Distribution of computed tomography (CT) scans without and with intracranial haemorrhage in the Prince of Wales Hospital dataset (n=1372)
Diagnostic performance of scan-based
detection for any type of intracranial haemorrhage
The model achieved an AUC of 0.842 (95% CI=0.791-0.894; P<0.001) for the identification of any type of ICH. Using a probability threshold of ≥50% for the
presence of ICH, the accuracy, sensitivity, specificity,
PPV, and NPV were 78.6%, 73%, 79%, 18.6%, and
97.8%, respectively. In total, 62 scans were true
positive, 22 were false negative, 1017 were true
negative, and 271 were false positive (Fig 2).
Among the 62 true-positive scans, the model
output in two cases did not contain ICH-positive CT
slices: 6-mm IPH in the pons (n=1) and trace SAH
in a patient with multiple metastatic tumours (n=1).
Figure 3 shows selected cases of model-nominated
CT slices with subtle ICH.
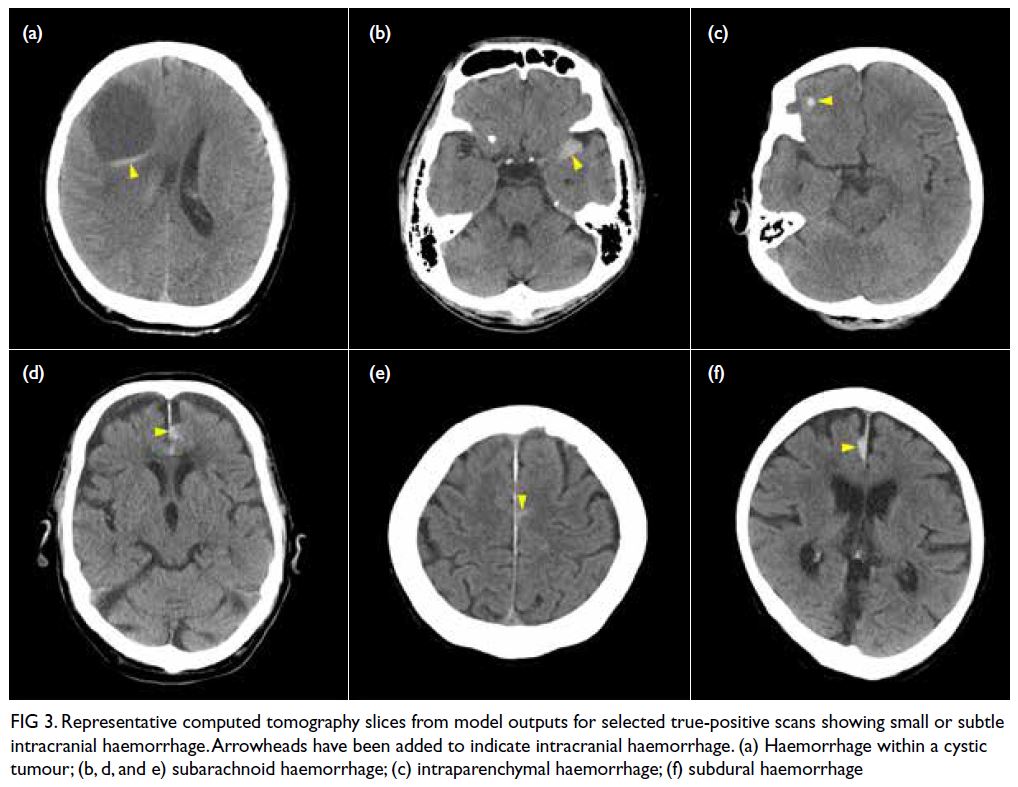
Figure 3. Representative computed tomography slices from model outputs for selected true-positive scans showing small or subtle intracranial haemorrhage. Arrowheads have been added to indicate intracranial haemorrhage. (a) Haemorrhage within a cystic tumour; (b, d, and e) subarachnoid haemorrhage; (c) intraparenchymal haemorrhage; (f) subdural haemorrhage
Among the 22 false-negative scans, 19 had one
type of ICH (6 IPH, 7 SAH, 5 SDH, and 1 IVH), two
had two types of ICH (1 IPH+SAH and 1 SAH+SDH),
and one had three types of ICH (IPH+SAH+IVH). In
16 scans, the model selected at least one ICH-positive
CT slice which allowed correct reclassification (Fig 4). The remaining six scans with undetected ICH
(Fig 5) comprised small midbrain IPH (n=1), trace
SAH (n=3), and thin SDH/EDH (n=2). One of the
three cases of undetected trace SAH was visualised
on thin CT slices but not on thick CT slices.
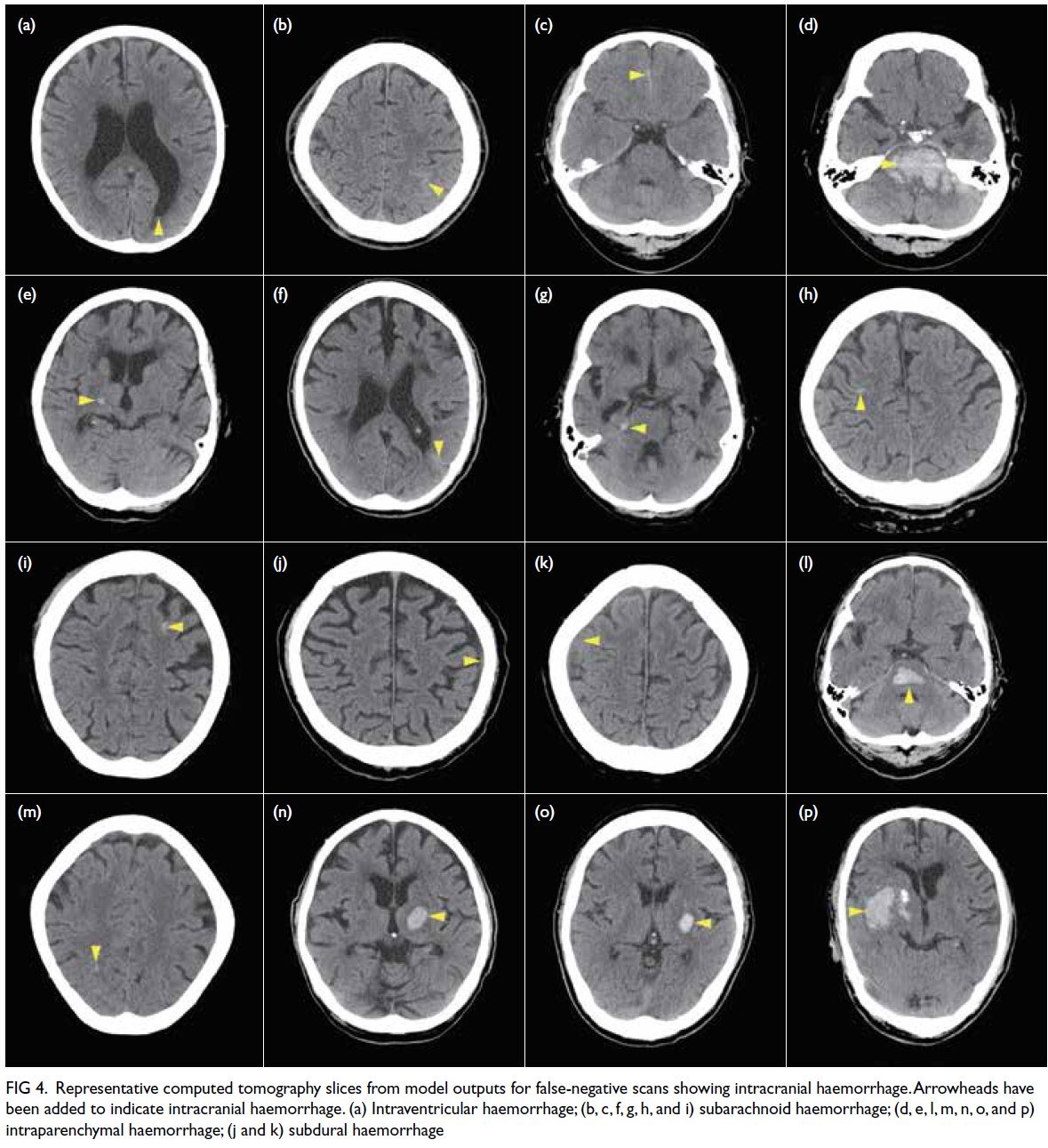
Figure 4. Representative computed tomography slices from model outputs for false-negative scans showing intracranial haemorrhage. Arrowheads have been added to indicate intracranial haemorrhage. (a) Intraventricular haemorrhage; (b, c, f, g, h, and i) subarachnoid haemorrhage; (d, e, l, m, n, o, and p) intraparenchymal haemorrhage; (j and k) subdural haemorrhage
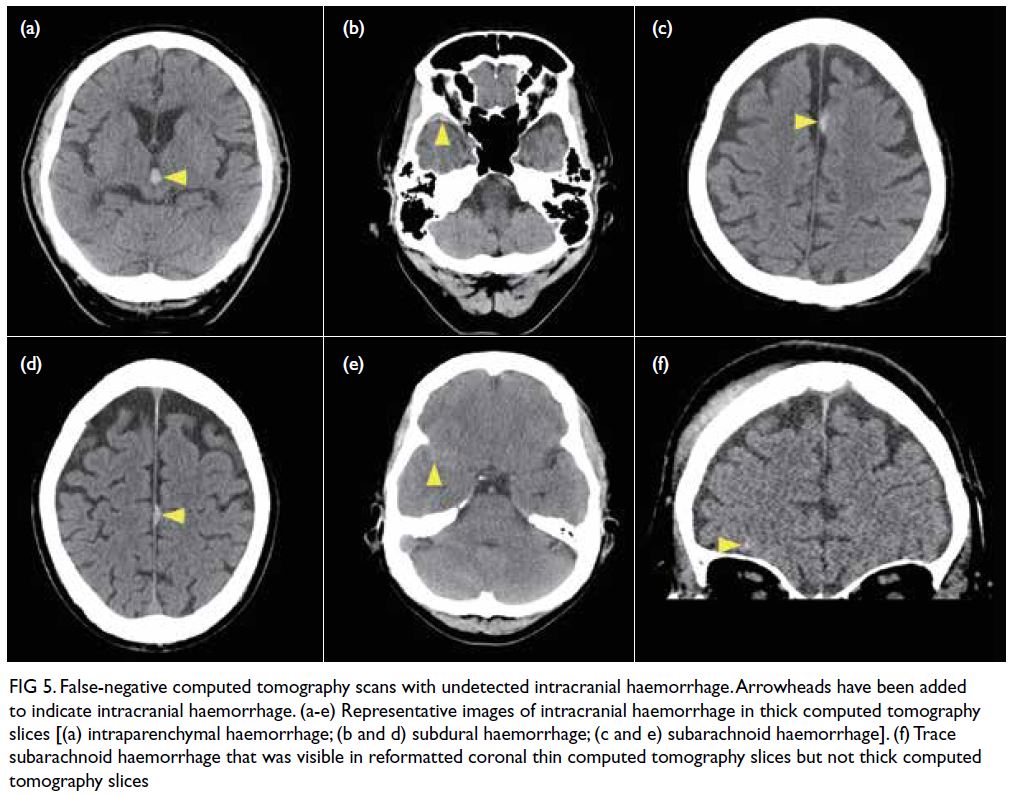
Figure 5. False-negative computed tomography scans with undetected intracranial haemorrhage. Arrowheads have been added to indicate intracranial haemorrhage. (a-e) Representative images of intracranial haemorrhage in thick computed tomography slices [(a) intraparenchymal haemorrhage; (b and d) subdural haemorrhage; (c and e) subarachnoid haemorrhage]. (f) Trace subarachnoid haemorrhage that was visible in reformatted coronal thin computed tomography slices but not thick computed tomography slices
A probability threshold of 20.4% yielded a
sensitivity of 90% (40% specificity, 9% PPV, and
98.3% NPV), whereas a threshold of 65.7% yielded
a specificity of 90% (64% sensitivity, 30% PPV, and
97.4% NPV), for the detection of ICH.
Diagnostic performance of scan-based
detection for each type of intracranial haemorrhage
At a probability threshold of ≥50%, the following
AUC (95% CI) and corresponding sensitivity/specificity were obtained for each type of ICH: 0.930 (0.892-0.968) and 4%/100% for IPH, 0.766 (0.684-0.849) and 12%/96% for SAH, 0.865 (0.783-0.947)
and 75%/90% for SDH/EDH, and 0.935 (0.852-1.000)
and 85%/93% for IVH.
Discussion
In this study, we used a large international training
dataset to construct a model for ICH detection,
then conducted external validation using data from
Hong Kong. To overcome the discrepancy between
the training dataset (composed of CT slices) and
the validation dataset (composed of CT scans),
and considering our goal of clinical application,
we designed a model that iteratively conducts
assessments at the slice level to generate an overall
probability at the scan level, then nominates the
slices with the highest ICH probability for clinician
evaluation. Furthermore, we performed validation
using a point-prevalence approach to determine the
diagnostic performance of the model in a real-world
setting. Considering the 6% prevalence of ICH in
our institution, and using a pre-specified probability
threshold of ≥50%, the model detected 74% of ICH-positive scans; this outcome improved to 93% via manual review of model-nominated images.
Artificial intelligence for intracranial
haemorrhage detection: research and reality
Multiple studies have successfully used AI
for ICH detection via deep learning methods,
typically involving variants of CNNs. For example,
Arbabshirani et al5 (deep CNN, >37 000 training CT
scans) reported an AUC of 0.846 on 342 CT scans;
Chang et al4 (two-dimensional/three-dimensional
CNN, 10 159 training CT scans) reported an AUC
of 0.983 on 862 prospectively collected CT scans. Furthermore, Chilamkurthy et al3 (CNN, >290 000
training CT scans) reported an AUC of 0.94 on 491
CT scans; Lee et al7 (four deep CNNs, 904 training
CT scans) reported an AUC of 0.96 on 214 CT
scans. Finally, Ye et al8 (three-dimensional joint
CNN-recurrent neural network, 2537 training CT
scans) reported an AUC of 1.0 on 299 CT scans;
Kuo et al6 (patch-based fully CNN, 4396 training
CT scans) reported an AUC of 0.991 on 200 CT
scans. Although these results demonstrate the
high diagnostic performance that can be achieved
using deep learning methods for ICH detection,
the studies were conducted using in-house training
datasets, which are laborious to produce and limit
subsequent clinical applications. Moreover, the
results may not be directly applicable to clinical
practice, considering the limited number (generally
<500) of CT scans during validation, as well as the
effect of prevalence on sensitivity and specificity.
Yune et al15 demonstrated this problem with a deep
learning model that had an AUC of 0.993 on selected
cases, which decreased to 0.834 when validated on
CT scans collected over a 3-month period; notably,
this is comparable with the AUC of our model.
Thus, model performance in a real-world setting can
reduce the risk of bias and serve as a better indicator
of clinical relevance.16
Artificial intelligence for intracranial
haemorrhage detection: our approach
The development of an AI model is the first step in a
long process of clinical translation. In this study, we
aimed to construct an algorithm that was reasonably
comparable with radiologist performance, prior to
further tests in a clinical setting. We recognise that
our model is not an end-product; it constitutes an
initial exploration of the potential for an international
dataset–derived algorithm to be implemented in our
institution. To avoid problems associated with the
lack of an annotated dataset from Hong Kong, we
utilised a dataset labelled by international experts,
which is the most extensive open-access dataset
currently available. However, the model achieved
limited diagnostic accuracy, mainly because of type
1 error (ie, identification of false positives). The
training dataset was composed of CT slices, whereas
the model functioned at the CT scan level, iteratively
assessing all slices to identify slices with highest ICH
probability. If any slice identified in a single scan is
considered positive, the model reports the CT scan as
‘ICH-positive’. Thus, any detection of false positives
at the slice level will lead to amplification of the false-positive
rate at the scan level. This strategy resulted in a low PPV (~19%) and a high NPV (~98%). To
reduce the detection of false positives, we included
a CT slice nomination feature in the model, which
highlights CT slices with the highest probability of
ICH. This facilitates manual review and reduces the
black-box nature of the model.
Potential implications of artificial
intelligence–detected intracranial haemorrhage in clinical practice
During validation, the model was tested using an
ICH point–prevalence approach to elucidate the
potential clinical implications of the classification
outcomes. With respect to true positives, most ICH-positive
scans were detected; most of these scans
had large areas of ICH, which presumably could be
easily identified by non-radiologists. However, in
six cases, the model correctly nominated CT slices
with small areas of ICH. In two cases, the nominated
images did not have ICH, which could potentially
have led to incorrect reclassification of the scan as
a false positive.
Furthermore, there were many false positives.
Such results may reduce physician confidence despite
the correct interpretation of an ICH-negative scan; they may lead to overdiagnosis (with prolonged
hospitalisation) or further investigations, such as a
follow-up CT scan that involves additional radiation
exposure.
With respect to false negatives, the model
output includes a secondary mechanism of image review that allowed correct reclassification of 16
scans, increasing the rate of ICH detection from
74% to 93%. In five cases, ICH was conspicuous on
the nominated images; in 11 cases, the nominated
images displayed subtle ICH. In cases of subtle ICH,
it is possible to overlook the trace amount of ICH on the nominated CT slice. The same problem may
affect true-positive scans, which may be misclassified
as false positives unless subtle ICH is recognised in
the nominated image. Unfortunately, the model-generated
probability of each type of ICH in each
selected image did not facilitate the localisation of ICH.
Based on our primary clinical motivation to
develop this model, we focused on CT scans with
reformatted thick CT slices that can be viewed
in all hospital workstations by non-radiologists.
In practice, radiologists use dedicated imaging
workstations to view sub-millimetre thin CT slices
with greater sensitivity, which can display smaller or
subtler pathologies. Thus, there is limited capacity
for ICH detection in thick CT slices; this was
highlighted in a case of trauma-related trace SAH,
which was visible on thin CT slices but not thick CT
slices. Subarachnoid haemorrhage is reportedly the
most difficult type of ICH to interpret.17 In practice,
a patient with a very small amount of isolated
traumatic SAH would likely receive conservative
treatment, and the pathology could reasonably await
detection via radiologist assessment.
Limitations
This study had some limitations. First, diagnostic
accuracy would have been more comprehensively
assessed using a larger number of CT scans or a
longer point prevalence; however, we limited the
assessment to CT scans collected over a 1-month
period, considering the preliminary stage of model
development. Second, the CT scans were assessed
by radiologists and senior radiology trainees who
may have different degrees of experience in ICH
detection17; importantly, this limitation reflects
the real-world setting where model deployment
is intended. Finally, the model was specifically
trained for the detection of ICH; it was not trained
for the detection of other clinically significant
non-ICH findings (eg, non-haemorrhagic tumours,
hydrocephalus, or mass effect). The detection of
these other pathologies will require dedicated
models with customised training datasets.
Conclusion
In this study, we used a CT slice–based dataset
to develop an algorithm for CT scan–based ICH detection; we validated the model using our
institutional data with a point-prevalence approach,
yielding insights regarding its utility in real-world
clinical practice. Although the model demonstrated
good accuracy, its diagnostic performance is
currently limited to the intended clinical application.
However, our results support further development of
the model to improve its accuracy and incorporate a
mechanism that can facilitate visual confirmation of
ICH location. These modifications would enhance
the interpretability of the deep learning model and
would be useful for further evaluation of clinical
applications.
Author contributions
Concept or design: JM Abrigo, KL Ko, WCW Chu, SCH Yu.
Acquisition of data: JM Abrigo, Q Chen, WCW Chu, BMH Lai, TCY Cheung.
Analysis or interpretation of data: All authors.
Drafting of the manuscript: JM Abrigo, KL Ko, Q Chen.
Critical revision of the manuscript for important intellectual content: All authors.
Acquisition of data: JM Abrigo, Q Chen, WCW Chu, BMH Lai, TCY Cheung.
Analysis or interpretation of data: All authors.
Drafting of the manuscript: JM Abrigo, KL Ko, Q Chen.
Critical revision of the manuscript for important intellectual content: All authors.
All authors had full access to the data, contributed to the study, approved the final version for publication, and take responsibility for its accuracy and integrity.
Conflicts of interest
All authors have disclosed no conflicts of interest.
Acknowledgement
We thank our department colleagues Mr Kevin Lo for
anonymising and downloading Digital Imaging and
Communications in Medicine data, and we thank Mr Kevin
Leung for preparing figures for this manuscript.
Funding/support
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Ethics approval
This research was approved by the Joint Chinese University of Hong Kong—New Territories East Cluster Clinical Research
Ethics Committee (Ref No.: 2020.061). The requirement for
patient consent was waived by the Committee given the
retrospective design of the study and anonymisation of all
computed tomography scans prior to use.
References
1. Caceres JA, Goldstein JN. Intracranial hemorrhage. Emerg Med Clin North Am 2012;30:771-94. Crossref
2. Powers WJ, Rabinstein AA, Ackerson T, et al. Guidelines
for the early management of patients with acute ischemic
stroke: 2019 update to the 2018 guidelines for the early
management of acute ischemic stroke: a guideline for
healthcare professionals from the American Heart
Association/American Stroke Association. Stroke 2019;50:e344-418. Crossref
3. Chilamkurthy S, Ghosh R, Tanamala S, et al. Deep learning algorithms for detection of critical findings in head CT
scans: a retrospective study. Lancet 2018;392:2388-96. Crossref
4. Chang PD, Kuoy E, Grinband J, et al. Hybrid 3D/2D convolutional neural network for hemorrhage evaluation
on head CT. AJNR Am J Neuroradiol 2018;39:1609-16. Crossref
5. Arbabshirani MR, Fornwalt BK, Mongelluzzo GJ, et al. Advanced machine learning in action: identification of
intracranial hemorrhage on computed tomography scans
of the head with clinical workflow integration. NPJ Digit
Med 2018;1:9. Crossref
6. Kuo W, Häne C, Mukherjee P, Malik J, Yuh EL. Expert-level
detection of acute intracranial hemorrhage on head
computed tomography using deep learning. Proc Natl
Acad Sci U S A 2019;116:22737-45. Crossref
7. Lee H, Yune S, Mansouri M, et al. An explainable deep-learning
algorithm for the detection of acute intracranial
haemorrhage from small datasets. Nat Biomed Eng 2019;3:173-82. Crossref
8. Ye H, Gao F, Yin Y, et al. Precise diagnosis of intracranial hemorrhage and subtypes using a three-dimensional joint convolutional and recurrent neural network. Eur Radiol
2019;29:6191-201. Crossref
9. Flanders AE, Prevedello LM, Shih G, et al. Construction of a machine learning dataset through collaboration: The
RSNA 2019 Brain CT Hemorrhage Challenge. Radiol Artif
Intell 2020;2:e190211. Crossref
10. Bossuyt PM, Reitsma JB, Bruns DE, et al. STARD 2015: an updated list of essential items for reporting diagnostic
accuracy studies. Radiology 2015;277:826-32. Crossref
11. Radiological Society of North America. RSNA intracranial hemorrhage detection: identify acute intracranial
hemorrhage and its subtypes. Available from: https://www.kaggle.com/c/rsna-intracranial-hemorrhage-detection/data. Accessed 28 Mar 2023. Crossref
12. Zhang X, Zou J, He K, Sun J. Accelerating very deep
convolutional networks for classification and detection.
IEEE Trans Pattern Anal Mach Intell 2016;38:1943-55. Crossref
13. Milletari F, Navab N, Ahmadi SA. V-Net: fully
convolutional neural networks for volumetric medical
image segmentation. 2016 Fourth International Conference
on 3D Vision (3DV). USA (CA): Stanford; 2016: 565-71. Crossref
14. DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating
characteristic curves: a nonparametric approach.
Biometrics 1988;44:837-45. Crossref
15. Yune S, Lee H, Pomerantz S, et al. Real-world performance
of deep-learning–based automated detection system for
intracranial hemorrhage. Radiological Society of North
America (RSNA) 104th Scientific Assembly and Annual
Meeting. McCormick Place, Chicago (IL); 2018.
16. Nagendran M, Chen Y, Lovejoy CA, et al. Artificial
intelligence versus clinicians: systematic review of design,
reporting standards, and claims of deep learning studies.
BMJ 2020;368:m689. Crossref
17. Strub WM, Leach JL, Tomsick T, Vagal A. Overnight preliminary head CT interpretations provided by residents:
locations of misidentified intracranial hemorrhage. AJNR
Am J Neuroradiol 2007;28:1679-82. Crossref