Hong Kong Med J 2015 Feb;21(1):16–22 | Epub 24 Oct 2014
DOI: 10.12809/hkmj144222
© Hong Kong Academy of Medicine. CC BY-NC-ND 4.0
ORIGINAL ARTICLE
Comparison between fluorescent in-situ hybridisation and array comparative genomic hybridisation in preimplantation genetic diagnosis in translocation carriers
Vivian CY Lee, FHKAM (Obstetrics and Gynaecology); Judy FC Chow, MPhil; Estella YL Lau, PhD; William SB Yeung, PhD; PC Ho, MD; Ernest HY Ng, MD
Department of Obstetrics and Gynaecology, The University of Hong Kong, Queen Mary Hospital, Pokfulam, Hong Kong
Corresponding author: Dr Vivian CY Lee (v200lee@hku.hk)
Abstract
Objectives: To compare the pregnancy outcome
of the fluorescent in-situ hybridisation and
array comparative genomic hybridisation in
preimplantation genetic diagnosis of translocation
carriers.
Design: Historical cohort.
Setting: A teaching hospital in Hong Kong.
Patients: All preimplantation genetic diagnosis
treatment cycles performed for translocation
carriers from 2001 to 2013.
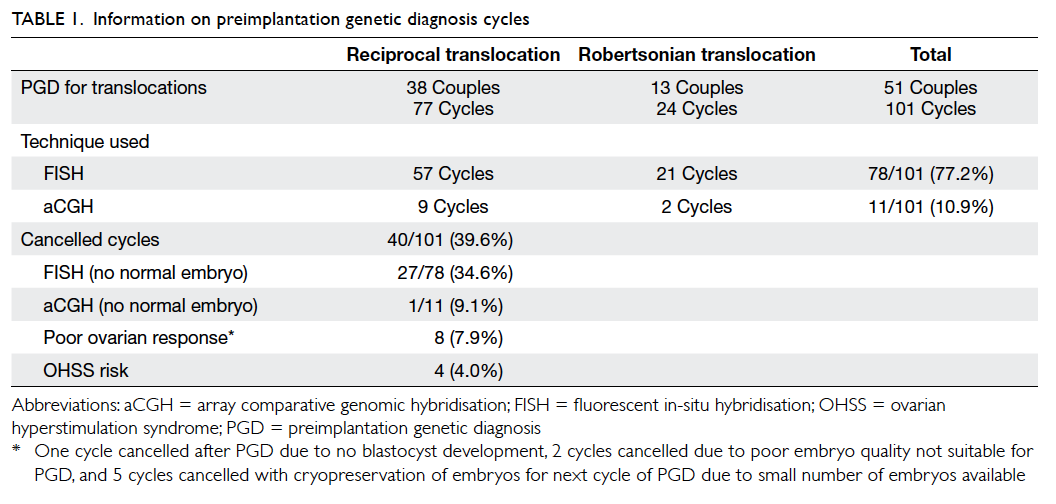
Results: Overall, 101 treatment cycles for
preimplantation genetic diagnosis in translocation
were included: 77 cycles for reciprocal translocation
and 24 cycles for Robertsonian translocation.
Fluorescent in-situ hybridisation and array
comparative genomic hybridisation were used in 78
and 11 cycles, respectively. The ongoing pregnancy
rate per initiated cycle after array comparative
genomic hybridisation was significantly higher
than that after fluorescent in-situ hybridisation
in all translocation carriers (36.4% vs 9.0%;
P=0.010). The miscarriage rate was comparable
with both techniques. The testing method (array
comparative genomic hybridisation or fluorescent
in-situ hybridisation) was the only significant
factor affecting the ongoing pregnancy rate after
controlling for the women’s age, type of translocation,
and clinical information of the preimplantation
genetic diagnosis cycles by logistic regression (odds
ratio=1.875; P=0.023; 95% confidence interval,
1.090-3.226).
Conclusion: This local retrospective study confirmed
that comparative genomic hybridisation is associated
with significantly higher pregnancy rates versus
fluorescent in-situ hybridisation in translocation
carriers. Array comparative genomic hybridisation
should be the technique of choice in preimplantation
genetic diagnosis cycles in translocation carriers.
New knowledge added by this
study
- Fluorescence in-situ hybridisation (FISH) has been widely used in preimplantation genetic diagnosis (PGD) in translocation carriers. However, array comparative genomic hybridisation (aCGH) has largely replaced FISH since its development due to the advantages of testing all 24 chromosomes and improved pregnancy rates. This is the first study to show the use of aCGH in Hong Kong. Compared with FISH, aCGH was associated with significantly higher rate of ongoing pregnancy in translocation carriers (both reciprocal and Robertsonian translocations).
- Array CGH should be the technique of choice for PGD in translocation carriers.
Introduction
Since the report of first live-birth after
preimplantation genetic diagnosis (PGD) published
in 1990,1 more than 21 000 cycles have been
performed worldwide, based on the data from
ESHRE (European Society of Human Reproduction
and Embryology) PGD consortium in the past two
decades.2 Fluorescent in-situ hybridisation (FISH)
has been used for PGD in translocation carriers. This
technique uses chromosome-specific DNA probes in
metaphase chromosomes or interphase nuclei. For
PGD in translocation carriers, the usual approach
is to use commercially available centromeric, locus-specific
and subtelomeric probes depending on the
translocated segments.3
However, FISH itself carries technical
difficulties of fixation and spreading of nucleus,
with the reported error rate of 7% to 10%.4 5 6 Another
problem is that in translocation carriers, there is
interchromosomal effect so that the proportion of
embryos having aneuploidies is higher than those
without translocations.7 Segmental loss or gain is also
a frequent event in human embryos.8 9 Fluorescent
in-situ hybridisation would not be able to detect
these chromosomal abnormalities, which could be
the cause of low success rates of PGD in translocation
carriers as most of these embryos would result in
implantation failure or miscarriages.10
With the development of comparative
genomic hybridisation (CGH), it is possible to
detect abnormalities in all 24 chromosomes and
its application on single blastomere biopsy was
first reported in 1996.11 Comparative genomic
hybridisation is a DNA-based technique, employing
comparative hybridisation of differentially labelled
DNA samples to normal metaphase chromosome on
a microscope slide.4 The ratio of fluorescence reveals
the gain or loss of the tested samples. However,
the turnover time is about 4 days, which does not
fit into the strict time frame of treatment for PGD,
and cryopreservation of embryos is mandatory,
unless polar body biopsy is used.12 Array CGH
(aCGH), employing DNA probes affixed directly
to a microscope slide, solves this problem as the
turnover time is about a day, which makes fresh
transfer after blastomere biopsy or trophectoderm
biopsy possible.13 It has been demonstrated that
using aCGH in translocation carriers is beneficial.9
Our centre used the FISH technique for
translocation carriers since our team developed the
technique of PGD in 2001 which resulted in the first
live-birth in Hong Kong.14 We acquired the platform
of aCGH in April 2012. This retrospective analysis
aimed to compare the pregnancy outcomes using
FISH and aCGH for the treatment cycles of PGD in
translocation carriers.
Methods
Study population
Data from all treatment cycles performed for PGD
in the Department of Obstetrics and Gynaecology,
Queen Mary Hospital/The University of Hong
Kong from 2001 till 2013 June were retrieved. Only
PGD cycles in translocation carriers were included
in the present study, which was approved by the
Institutional Review Board of the University of Hong
Kong/Hospital Authority Hong Kong West Cluster.
Treatment regimen
The details of the long protocol of ovarian stimulation
regimen, gamete handling, cryopreservation of
embryos, and frozen embryo transfer have been
previously described.15 The details of PGD have
also been previously described.16 In short, embryo
biopsy was performed on day 3 at 6-to-8-cell stage.
Two blastomeres were tested from 2001 to 2005
and one blastomere was routinely tested from 2006
onwards. The blastomere was fixed for FISH analysis.
Commercially available FISH probes were chosen to
flank the break point. For aCGH, the blastomere was
transferred into a polymerase chain reaction tube
and whole genome amplification was performed
(SurePlex; BlueGnome, Cambridge, UK). Array
CGH was performed using 24sure V3 (BlueGnome)
for Robertsonian translocation carrier or 24sure+
(BlueGnome) for reciprocal translocation carrier. All
results were interpreted separately by two laboratory
staff.
Outcome measures
The primary outcome measures of the study were
clinical and ongoing pregnancy rates. Clinical
pregnancies were defined by the presence of
one or more gestation sacs or the histological
confirmation of gestational product in case of early
pregnancy failures. Ongoing pregnancies were those
pregnancies beyond 8 to 10 weeks of gestation, at
which stage the patients were referred for antenatal
care. The secondary outcome measures were
miscarriage rate and cancellation rate. Cancellation
rate was defined as the percentage of treatment
cycles with no embryo transfer after oocyte retrieval.
Statistical analysis
The Kolmogorov-Smirnov test was used to test
the normal distribution of continuous variables.
Results of continuous variables were expressed as
mean ± standard deviation if normally distributed,
and median (range) if not normally distributed.
Statistical comparison was carried out by Student’s t
test, Mann-Whitney U test, and/or Wilcoxon signed
rank test for continuous variables and Chi squared
test or Fisher’s exact test for categorical variables,
as appropriate. Statistical analysis was performed
using the Statistical Package for the Social Sciences
(Windows version 20.0; SPSS Inc, Chicago [IL],
US). The two-tailed value of P<0.05 was considered
statistically significant. Binary logistic regression
using enter method was used to calculate the
prediction of the pregnancy rate in PGD cycles.
Results
There were 339 PGD cycles, of which 101 treatment
cycles were performed in translocation carriers
during the study period: 77 cycles for reciprocal
translocation and 24 cycles for Robertsonian
translocation. The two techniques, FISH and aCGH,
were used in 78 and 11 cycles, respectively (Table 1). The overall cancellation rate was 39.6% (40/101). Four cycles were cancelled due
to high risk of ovarian hyperstimulation syndrome;
eight cycles due to poor ovarian responses or poor
embryo qualities; and 28 cycles due to no normal
embryo after PGD with either technique (Table 1). The cancellation rate using FISH technique due to
abnormal signals for all embryos was significantly
higher than that using aCGH (34.6% vs 9.1%,
respectively).
The demographic and clinical data of women
who underwent PGD with FISH and aCGH are
presented in Table 2. Women in the aCGH group
were significantly younger than those in the FISH
group, and the serum oestradiol concentration
on ovulation trigger day in the aCGH group was
significantly higher than that in the FISH group.
The total dosage of gonadotropin, the number of
follicles larger than or equal to 16 mm, and the number of
oocytes retrieved were comparable between the
two groups. The demographic and clinical data of
cycles for couples with reciprocal and Robertsonian
translocation were all comparable (data not shown).
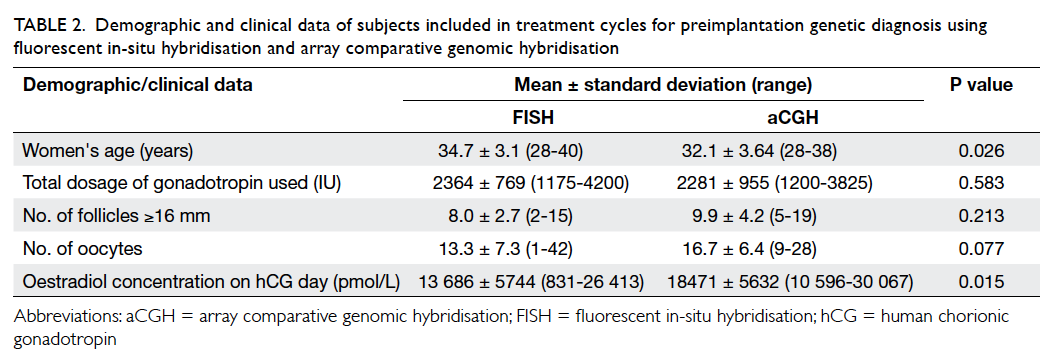
Table 2. Demographic and clinical data of subjects included in treatment cycles for preimplantation genetic diagnosis using fluorescent in-situ hybridisation and array comparative genomic hybridisation
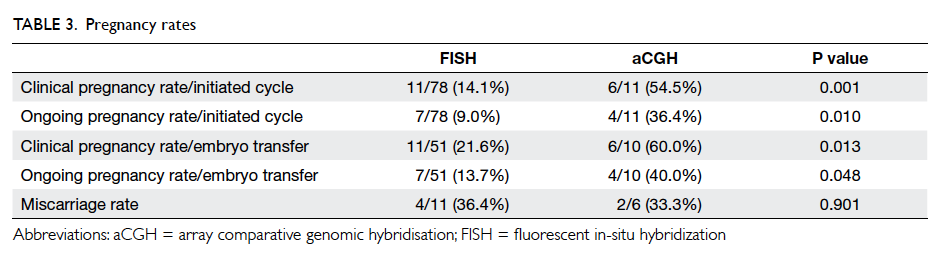
The pregnancy rates per cycle and per transfer
were all significantly higher in cycles performed
using aCGH. The miscarriage rates were similar
between the two groups (Table 3). A subgroup
analysis of cycles performed from 2006 to 2013
showed similar results in all the above comparisons
with significantly higher clinical and ongoing
pregnancy rates per initiated cycle and per transfer
in cycles using aCGH than those using FISH, but
with comparable miscarriage rates (data not shown).
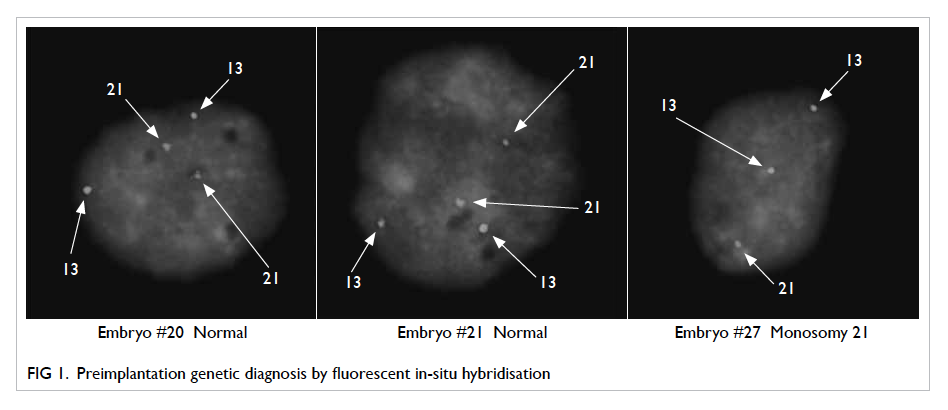
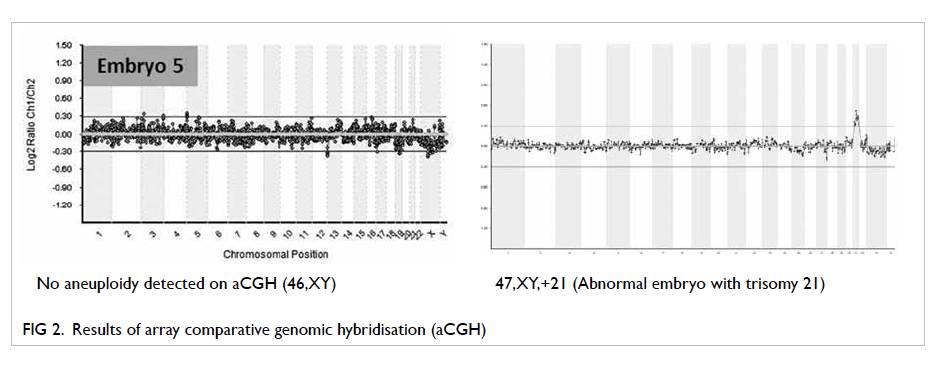
Figures 1 and 2 show PGD results with FISH and
aCGH, respectively.
Logistic regression revealed that the method
of testing (FISH or aCGH) was the only factor that
significantly affected the ongoing pregnancy rate;
age of the women, the type of translocation, or other
clinical information including number of oocytes
retrieved, the gonadotropin dosage used, and the
oestradiol concentration on the day of human
chorionic gonadotropin administration did not
affect the outcome. The method of testing remained
a significant factor after controlling for the age of women
and type of translocation (Table 4).
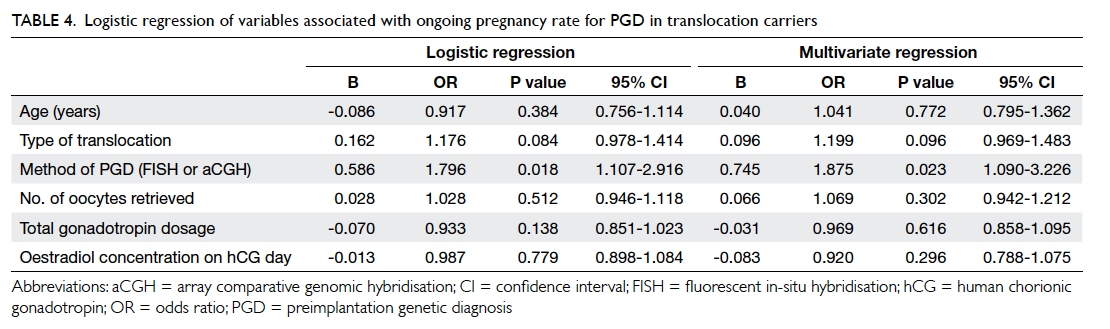
Table 4. Logistic regression of variables associated with ongoing pregnancy rate for PGD in translocation carriers
Figure 3 shows the results of aCGH in
embryos produced from reciprocal translocation
carrier. Array CGH can detect segmental changes in
translocated chromosomes (embryo 18) and other
chromosomes (embryo 3). It can also detect whole
chromosome aneuploidy (embryo 16). Embryo 7 was
replaced and resulted in an ongoing pregnancy.
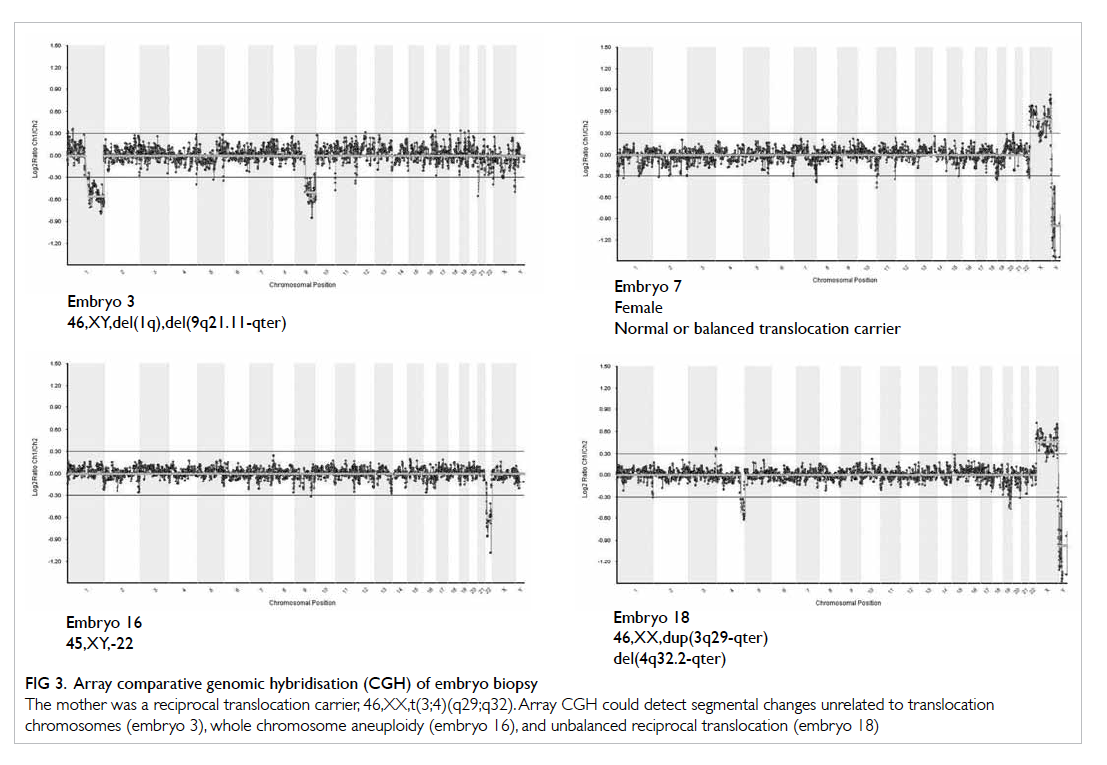
Figure 3. Array comparative genomic hybridisation (CGH) of embryo biopsy
The mother was a reciprocal translocation carrier, 46,XX,t(3;4)(q29;q32). Array CGH could detect segmental changes unrelated to translocation chromosomes (embryo 3), whole chromosome aneuploidy (embryo 16), and unbalanced reciprocal translocation (embryo 18)
Discussion
The present study showed that PGD using aCGH was
associated with significantly higher pregnancy rates
(both per initiated cycle or per embryo transfer)
versus FISH. The testing method, ie using aCGH or
FISH, was the only significant factor affecting the
ongoing pregnancy rate in logistic regression.
Couples carrying balanced reciprocal or
Robertsonian translocations are well-known to
produce a high percentage of unbalanced gametes
and embryos,17 resulting in high miscarriage rates
and a variable chance of unbalanced offspring
with multiple congenital anomalies and mental
retardation.18 The high percentage of unbalanced
gametes can be explained by the segregation modes
and behaviours of translocations during meiosis.19
Not only the direct effect of the translocations
on the meiosis, but also the interchromosomal
effect exerted by the translocations increases the
percentage of aneuploidies in the gametes and
embryos of couples carrying translocations.7 20 21 22 It
further decreases the number of normal/balanced
euploid embryos, including those suitable and
feasible for transfer. It was reported that only up
to 16% of preimplantation embryos were normal/balanced and euploid in translocation carriers.9
In the past decade, FISH was commonly
employed to detect the unbalanced chromosome
rearrangement of embryos using probes depending
on the translocated segments.23 Fluorescent
in-situ hybridisation is technically challenging,
especially with regard to fixation and spreading.3 5 24
The error rate of FISH was reported to be up to
10%.5 6 25 As PGD using FISH in translocation carriers only employs fluorescent DNA probes
for the translocated segments, aneuploidies and
segmental rearrangements which are not related to
the translocated segments will be missed.3 Even in
aneuploidy screening, only up to five chromosomes
can be tested in one round of FISH, and so, usually
up to half of all chromosomes can be tested in
repeated rounds. However, repeated rounds were
related to the decrease in diagnostic accuracy.4
Therefore, using FISH would miss a proportion of
aneuploidies and abnormal embryos, which may
result in misdiagnosis, implantation failure, or
miscarriages.10 This is probably the major reason for
the unfavourable results in a systematic review on the
use of PGD in translocation carriers26 and the meta-analysis
of preimplantation genetic screening.27 The
cancellation rate, ie no embryo transfer after oocyte
retrieval, was higher after FISH than that after
aCGH, probably due to technical difficulties.
The development of CGH makes it possible to
test for all 24 chromosomes, while the development
of aCGH makes it feasible to use the technique in
the restricted time frame of PGD. Several groups
of investigators have reported success with using
aCGH for PGD in translocation carrier couples to
improve their reproductive outcomes9 28; we have
shown similar results in this local study.
Figure 3 shows the result of PGD in a patient with reciprocal translocation. Array CGH detected
unbalanced reciprocal translocated segments in
embryo 18. It also picked other segmental changes
(1q and 9q21.11-qter) not related to translocated
chromosomes in embryo 3. It could also detect
whole chromosome aneuploidy (monosomy
22) in embryo 16. In FISH, probes flanking the
translocation breakpoints are used and, therefore, the
abnormalities in embryo 3 and embryo 16 cannot be
detected. Furthermore, the average probe density of
aCGH used for Robertsonian translocation is 10 Mb
while that of one used for reciprocal translocation is
5 Mb. Increase in resolution allows us to easily pick a
small abnormality in the embryo. Array CGH offers
a more comprehensive way of PGD in translocation
carriers and this results in a significant increase in
the pregnancy rate compared with FISH.
In our cohort, the age of women for whom
aCGH was employed was younger than that of
women for whom FISH was employed. This can
probably explain the higher oestradiol concentration
after ovarian stimulation of in-vitro fertilisation
treatment, along with the non-significant, higher
number of follicles and oocytes retrieved in the
aCGH group. In order to reveal the effect of the
testing method on pregnancy rate, we controlled
the women’s age, type of translocation, and other
data of the stimulation including the total dosage of
gonadotropin and number of oocytes retrieved in
multivariate logistic regression; the testing method
remained the only significant factor affecting the
ongoing pregnancy rate. This indicates that, after
controlling for all the possible confounding factors,
PGD cycles using aCGH were associated with a
significantly higher ongoing pregnancy rate than
those using FISH.
It has been controversial whether PGD can
improve the reproductive outcomes compared
with natural conception in translocation carriers.
A systematic review reported adverse effects on the
pregnancy rates after PGD in translocation carriers
compared with natural conception.26 However, all the
PGD cycles included in this review were performed
with FISH. Moreover, the case reports and case series
of PGD included had a small number of subjects; in
16 out of 21 studies, the sample size was only one
to three cases. Larger systematic reviews on the use
of aCGH in translocation carriers are urgently
needed.
This study is retrospective in nature and there
may be some confounding factors such as differences
in embryo biopsy techniques and culture conditions
which were not controlled for and which might have
affected the pregnancy outcomes. As we started
using aCGH approximately one and a half year ago,
the number of cases was smaller than that using
FISH. Despite the small sample size, the ongoing
pregnancy rate revealed a significant increase after
employing aCGH in translocation carriers. This
serves to further strengthen our argument in favour
of PGD programme using aCGH.
It is well known that two-blastomere biopsy is
more detrimental to pregnancy than one-blastomere
biopsy.22 Our team employed two-blastomere biopsy
when we first developed our PGD programme. We
then switched to one-blastomere biopsy in 2006.
Therefore, a subgroup analysis was performed on
those cycles between 2006 and 2013. The ongoing
pregnancy rate per initiated cycle remained
significantly higher in the group using aCGH than
that using FISH.
Conclusion
Use of aCGH can improve the pregnancy outcomes
of PGD in translocation carriers compared with
FISH. Array CGH should be the technique of choice
for PGD in translocation carriers.
References
1. Handyside AH, Kontogianni EH, Hardy K, Winston
R. Pregnancies from biopsied human preimplantation
embryos sexed by Y-specific DNA amplification. Nature
1990;344:768-70. CrossRef
2. Traeger-Synodinos J, Coonen E, Goossens V, et al. Session
09: ESHRE data reporting on PGD cycles and oocyte
donation. Hum Reprod 2013;28(Suppl 1):i18-i19. CrossRef
3. DeUgarte CM, Li M, Surrey M, Danzer H, Hill D, DeCherney
AH. Accuracy of FISH analysis in predicting chromosomal
status in patients undergoing preimplantation genetic
diagnosis. Fertil Steril 2008;90:1049-54. CrossRef
4. Wells D, Alfarawati S, Fragouli E. Use of comprehensive
chromosomal screening for embryo assessment:
microarrays and CGH. Mol Hum Reprod 2008;14:703-10. CrossRef
5. Velilla E, Escudero T, Munné S. Blastomere fixation
techniques and risk of misdiagnosis for preimplantation
genetic diagnosis of aneuploidy. Reprod Biomed Online
2002;4:210-7. CrossRef
6. Li M, DeUgarte CM, Surrey M, Danzer H, DeCherney A,
Hill DL. Fluorescence in situ hybridization reanalysis of
day-6 human blastocysts diagnosed with aneuploidy on
day 3. Fertil Steril 2005;84:1395-400. CrossRef
7. Gianaroli L, Magli MC, Ferraretti AP, et al. Possible
interchromosomal effect in embryos generated by gametes
from translocation carriers. Hum Reprod 2002;17:3201-7. CrossRef
8. Vanneste E, Voet T, Le Caignec C, et al. Chromosome
instability is common in human cleavage-stage embryos. CrossRef
Nat Med 2009;15:577-83.
9. Fiorentino F, Spizzichino L, Bono S, et al. PGD for reciprocal
and Robertsonian translocations using array comparative
genomic hybridization. Hum Reprod 2011;26:1925-35. CrossRef
10. Scott RT Jr, Ferry K, Su J, Tao X, Scott K, Treff NR.
Comprehensive chromosome screening is highly
predictive of the reproductive potential of human embryos:
a prospective, blinded, nonselection study. Fertil Steril
2012;97:870-5. CrossRef
11. Wells D, Delhanty J. Evaluating comparative genomic
hybridisation (CGH) as a strategy for preimplantation
diagnosis of unbalanced chromosome complements. Eur J
Hum Genet 1996;4:125.
12. Wells D, Escudero T, Levy B, Hirschhorn K, Delhanty JD,
Munné S. First clinical application of comparative genomic
hybridization and polar body testing for preimplantation
genetic diagnosis of aneuploidy. Fertil Steril 2002;78:543-9. CrossRef
13. Rubio C, Rodrigo L, Mir P, et al. Use of array comparative
genomic hybridization (array-CGH) for embryo
assessment: clinical results. Fertil Steril 2013;99:1044-8. CrossRef
14. Ng EH, Lau EY, Yeung WS, Lau ET, Tang MH, Ho PC. Preimplantation
genetic diagnosis in Hong Kong. Hong Kong
Med J 2003;9:43-7.
15. Ng EH, Yeung WS, Lau EY, So WW, Ho PC. High serum
oestradiol concentrations in fresh IVF cycles do not impair
implantation and pregnancy rates in subsequent frozen-thawed
embryo transfer cycles. Hum Reprod 2000;15:250-5. CrossRef
16. Chow JF, Yeung WS, Lau EY, et al. Singleton birth after
preimplantation genetic diagnosis for Huntington disease
using whole genome amplification. Fertil Steril 2009;92:828.e7-10.
17. Munné S. Analysis of chromosome segregation during
preimplantation genetic diagnosis in both male and female
translocation heterozygotes. Cytogenet Genome Res
2005;111:305-9. CrossRef
18. Jalbert P, Sele B, Jalbert H. Reciprocal translocations: a
way to predict the mode of imbalanced segregation by
pachytene-diagram drawing. Hum Genet 1980;55:209-22. CrossRef
19. Scriven PN, Handyside AH, Ogilvie CM. Chromosome
translocations: segregation modes and strategies
for preimplantation genetic diagnosis. Prenat Diagn
1998;18:1437-49. CrossRef
20. Pellestor F, Imbert I, Andréo B, Lefort G. Study of the
occurrence of interchromosomal effect in spermatozoa
of chromosomal rearrangement carriers by fluorescence
in-situ hybridization and primed in-situ labelling
techniques. Hum Reprod 2001;16:1155-64. CrossRef
21. Douet-Guilbert N, Bris MJ, Amice V, et al. Interchromosomal
effect in sperm of males with translocations:
report of 6 cases and review of the literature. Int J Androl
2005;28:372-9. CrossRef
22. Machev N, Gosset P, Warter S, Treger M, Schillinger M,
Viville S. Fluorescence in situ hybridization sperm analysis
of six translocation carriers provides evidence of an
interchromosomal effect. Fertil Steril 2005;84:365-73. CrossRef
23. Harper JC, Wilton L, Traeger-Synodinos J, et al. The
ESHRE PGD Consortium: 10 years of data collection. Hum
Reprod Update 2012;18:234-47. CrossRef
24. Munné S. Preimplantation genetic diagnosis of numerical
and structural chromosome abnormalities. Reprod Biomed
Online 2002;4:183-96. CrossRef
25. Munné S, Sandalinas M, Escudero T, Fung J, Gianaroli L,
Cohen J. Outcome of preimplantation genetic diagnosis of
translocations. Fertil Steril 2000;73:1209-18. CrossRef
26. Franssen MT, Musters AM, van der Veen F, et al.
Reproductive outcome after PGD in couples with
recurrent miscarriage carrying a structural chromosome
abnormality: a systematic review. Hum Reprod Update
2011;17:467-75. CrossRef
27. Mastenbroek S, Twisk M, van der Veen F, Repping S.
Preimplantation genetic screening: a systematic review and
meta-analysis of RCTs. Hum Reprod Update 2011;17:454-66. CrossRef
28. Colls P, Escudero T, Fischer J, et al. Validation of array
comparative genome hybridization for diagnosis of
translocations in preimplantation human embryos. Reprod
Biomed Online 2012;24:621-9. CrossRef
Find HKMJ in MEDLINE: