© Hong Kong Academy of Medicine. CC BY-NC-ND 4.0
CASE REPORT
Grave impact of undetected rpoB I572F mutation
on clinical course of multidrug-resistant tuberculosis: a case report
Alan CK Chan, MRCP (UK), FHKAM (Medicine)1; Martin CH Chan, MB, BS2; Peter CW Yip, PhD2; WC Yam, PhD, FRCPath3; CH Chau, MRCP, FHKAM (Medicine)4; Raymond FM Lam, MB, ChB, FHKCP4; LB Tai, MRCP, FHKAM (Medicine)1; CC Leung, FFPH, FHKAM (Medicine)5
1 Tuberculosis and Chest Service, Department of Health, Hong Kong SAR Government, Hong Kong
2 Public Health Laboratory Service Branch, Department of Health, Hong Kong SAR Government, Hong Kong
3 Department of Microbiology, Queen Mary Hospital, The University of Hong Kong, Hong Kong
4 Tuberculosis and Chest Unit, Grantham Hospital, Hong Kong
5 Hong Kong Tuberculosis, Chest and Heart Diseases Association, Hong Kong
Corresponding author: Dr Alan CK Chan (chikuen_chan@dh.gov.hk)
Case report
Accelerating diagnosis and treatment of rifampicin-resistant/multidrug-resistant tuberculosis (MDR-TB)
are key components of the World Health
Organization’s End TB Strategy.1 Rifampicin
resistance that arises from mutations outside the
81-base pair (bp) rifampicin-resistance determining
region (RRDR) of the rpoB gene such as the I572F
mutation nonetheless cannot be detected by
existing World Health Organization–endorsed
Xpert MTB/RIF assay or line probe assays (LPAs).2
Mutations located outside the rpoB hotspot
may also be missed by the liquid medium–based
BACTEC Mycobacteria Growth Indicator Tube
(MGIT) culture system. We report a rare case of
MDR-TB with rpoB I572F mutation (Escherichia
coli numbering system) that was missed by LPA
and liquid culture but confirmed by full rpoB gene
sequencing to illustrate the negative impact of the
mutation being undetected.
A 54-year-old Chinese man presented with
symptoms of TB in July 2019. He had no history of
anti-TB treatment. Chest radiograph on presentation
showed bilateral cavitary lesions (Fig a). Sputum
acid-fast bacilli smear examination was positive.
Xpert MTB/RIF assay confirmed Mycobacterium
tuberculosis (MTB)–positive/rifampicin-negative
pulmonary TB. He was prescribed standard anti-TB
treatment with isoniazid, rifampicin, ethambutol,
and pyrazinamide. Initial sputum culture (performed
with an MGIT culture system) later confirmed
MTB organisms susceptible to all first-line drugs.
Nonetheless lung shadows did not improve on
serial chest radiographs (Fig b and c) despite good
compliance with directly observed therapy. Sputum
culture was transiently negative between November
2019 and January 2020. Culture result of a sputum
specimen saved in October 2019 became available in January 2020 and showed MTB organisms resistant
to isoniazid. The LPA (GenoType MTBDRplus
Version 2.0; Hain Lifescience GmbH, Nehren,
Germany) performed at that juncture showed an
inhA C-15T mutation but no mutation in the rpoB
gene. The regimen was switched to rifampicin,
levofloxacin, ethambutol and pyrazinamide. Sputum
culture reverted to positive in subsequent months
and acid-fast bacilli smear also reverted to positive at
12 months. The LPA repeated at 12 months showed
inhA C-15T and gyrA D94A mutations. Mutation of
rpoB, rrs and enhanced intracellular survival genes
was not detected although whole genome sequencing
(WGS) performed on isolates obtained at 12 months
revealed mutations of rpoB I572F, inhA C-15T, embB
D354A, pncA D63G, and gyrA D94A. Whole genome
sequencing performed retrospectively on isolates
obtained earlier confirmed that rpoB I572F and
inhA C-15T mutations had been present since the
beginning of treatment. Culture result (using liquid
culture system) of a sputum specimen saved at 12
months later became available and showed bacillary
resistance to streptomycin, isoniazid, ethambutol
and levofloxacin, but rifampicin resistance was again
missed. In view of the gene sequencing result, the
regimen was switched to a bedaquiline-containing
regimen at 12 months. The patient responded well to
treatment thereafter (Fig d).
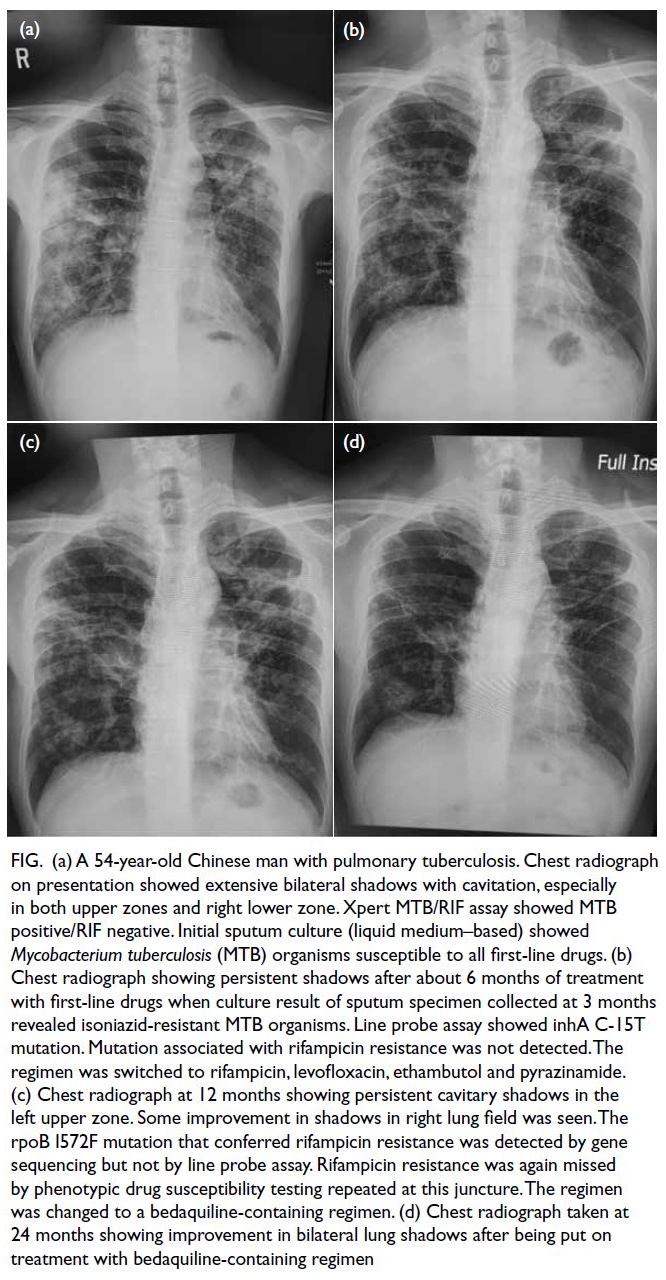
Figure. (a) A 54-year-old Chinese man with pulmonary tuberculosis. Chest radiograph on presentation showed extensive bilateral shadows with cavitation, especially in both upper zones and right lower zone. Xpert MTB/RIF assay showed MTB positive/RIF negative. Initial sputum culture (liquid medium–based) showed Mycobacterium tuberculosis (MTB) organisms susceptible to all first-line drugs. (b) Chest radiograph showing persistent shadows after about 6 months of treatment with first-line drugs when culture result of sputum specimen collected at 3 months revealed isoniazid-resistant MTB organisms. Line probe assay showed inhA C-15T mutation. Mutation associated with rifampicin resistance was not detected. The regimen was switched to rifampicin, levofloxacin, ethambutol and pyrazinamide. (c) Chest radiograph at 12 months showing persistent cavitary shadows in the left upper zone. Some improvement in shadows in right lung field was seen. The rpoB I572F mutation that conferred rifampicin resistance was detected by gene sequencing but not by line probe assay. Rifampicin resistance was again missed by phenotypic drug susceptibility testing repeated at this juncture. The regimen was changed to a bedaquiline-containing regimen. (d) Chest radiograph taken at 24 months showing improvement in bilateral lung shadows after being put on treatment with bedaquiline-containing regimen
Discussion
Although most cases of rifampicin resistance are
linked to mutations in the 81-bp hotspot region of the
rpoB gene, notably mutations at codons 526 to 531,
our case illustrates the occurrence of rare mutations
outside the RRDR, namely I572F, and its negative
impact on treatment outcome due to amplification
of further drug resistance if left undetected. In our
case, initial rifampicin resistance conferred by rpoB I572F mutation was missed by Xpert MTB/RIF
assay, LPA and liquid medium–based culture system.
Due to the exceptionally slow growth of this strain,
prolonged incubation using MGIT was required and
there was also difficulty in selecting log phase growth
for subsequent drug susceptibility testing (DST).
Not only was turnaround time increased, but initial
isoniazid resistance was also missed although it was
detected in the subsequent strains phenotypically.
The patient therefore received an inadequate number
of drugs during the early course of anti-TB treatment
resulting in further acquired drug resistance to
ethambutol, pyrazinamide, and levofloxacin.
Amplification of drug resistance in our case likely
emerged from the segregation of a single strain into
two lineages of drug-susceptible and drug-resistant
organisms under the selective pressure of insufficient
TB therapy. This was suggested by the presence of
both wild type and resistant subpopulations during
the transition from susceptibility to resistance with
regard to levofloxacin from the WGS data on D94A
mutation.
Review of the database at the TB Supranational
Reference Laboratory, Centre for Health Protection,
Department of Health of Hong Kong revealed a
total of five cases of MDR-TB with rpoB I572F
mutation (including the present case) out of 340
rifampicin-resistant isolates between 2011 and
2020, corresponding to a prevalence of 1.5%. The
prevalence of rpoB I572F in Hong Kong in this study
is similar to that (2%) reported from a previous local
study.3 On the contrary, a much higher prevalence
of rpoB I572F mutation (corresponding to Ile491Phe
mutation in MTB numbering system) has been
reported recently in some countries with high TB
prevalence such as Eswatini (formerly Swaziland)
and South Africa (30% and 15%, respectively).4 The
highly variable prevalence of rpoB I572F mutation
in different geographical regions highlights the
importance of expanding the geographical database
of this mutation to better understand its global
prevalence.
To improve the accuracy of phenotypic
DST, the World Health Organization has recently
lowered the critical concentration for rifampicin
susceptibility testing in MGIT from 1 mg/L to
0.5 mg/L.5 The revised recommendation helps
reduce but does not eliminate the discordance
observed between phenotypic and molecular
methods to detect rifampicin resistance and the
potential false-susceptible results from phenotypic
tests due to the presence of mutations outside the
RRDR. Given the potential impact of rifampicin
resistance conferred by an rpoB I572F mutation on
treatment outcomes, and that an increasingly higher
prevalence of such mutations has been reported
recently in some countries, new molecular tests that
expand the drug target coverage to help guide the formulation of treatment regimens are warranted.
Expanding the target 81-bp hotspot RRDR of the
rpoB gene to include codon 572 has been suggested.3
More recently, a multiplex allele-specific polymerase
chain reaction (PCR) assay to detect I572F mutation
in rpoB has been designed using a one-step real-time
PCR.6 Although novel PCR assays may enable
efficient and rapid detection of rpoB I572F mutation and are simpler than sequencing methods, their
implementation requires further validation and
strengthening of laboratory capacities.
Until novel PCR assays and WGS gain
widespread use, clinicians should remain alert
for the more rare rpoB mutations such as the
I572F mutation, and communicate promptly with
laboratories for further tests if a patient does not
respond well to standard first-line treatment.
This is vital even if Xpert MTB/RIF assay, LPAs
or phenotypic DST do not suggest the presence of
rifampicin resistance to ensure the patient receives
an adequate number of effective drugs for treatment
success. Our case also calls for continued surveillance
of the prevalence of rpoB I572F mutation and other
rifampicin-resistance conferring mutations outside
the RRDR to inform region-specific TB diagnostic
and treatment strategies.
Author contributions
All authors contributed to the concept or design of the study, acquisition of the data, analysis or interpretation of the
data, drafting of the manuscript, and critical revision of the
manuscript for important intellectual content.
All authors had full access to the data, contributed to the study, approved the final version for publication, and take responsibility for its accuracy and integrity.
Conflicts of interest
The authors have disclosed no conflicts of interest.
Declaration
Part of the findings of this study has been presented at the Hong Kong Thoracic Society clinical meeting on 26 May 2022, which was an internal meeting attended by members of the
Society held in virtual format.
Funding/support
This study received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Ethics approval
The patient was treated in accordance with the Declaration of Helsinki and provided written informed consent for
publication.
References
1. World Health Organization. The end TB strategy. 2015. Available from: https://www.who.int/publications/i/item/WHO-HTM-TB-2015.19. Accessed 31 Jan 2023.
2. Nguyen TN, Anton-Le Berre V, Bañuls AL, Nguyen TV. Molecular diagnosis of drug-resistant tuberculosis; a
literature review. Front Microbiol 2019;10:794. Crossref
3. Siu GK, Zhang Y, Lau TC, et al. Mutations outside the rifampicin resistance-determining region associated with rifampicin resistance in Mycobacterium tuberculosis. J
Antimicrob Chemother 2011;66:730-3. Crossref
4. Variava E, Martinson N. Occult rifampicin-resistant
tuberculosis: better assays are needed. Lancet Infect Dis
2018;18:1293-5. Crossref
5. World Health Organization. Technical report on critical
concentrations for drug susceptibility testing of isoniazid
and the rifamycins (rifampicin, rifabutin and rifapentine).
Geneva: World Health Organization; 2021.
6. André E, Goeminne L, Colmant A, Beckert P, Niemann S, Delmee M. Novel rapid PCR for the detection of
Ile491Phe rpoB mutation of Mycobacterium tuberculosis, a
rifampicin-resistance-conferring mutation undetected by
commercial assays. Clin Microbiol Infect 2017;23:267.e5-7. Crossref

